hnn_core.jones_2009_model#
- hnn_core.jones_2009_model(params=None, add_drives_from_params=False, legacy_mode=False, mesh_shape=(10, 10))[source]#
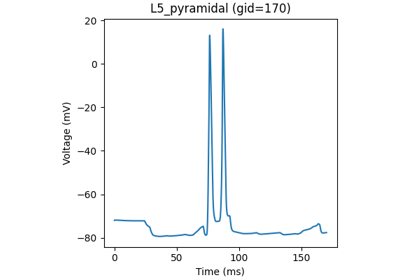
Instantiate the network model described in Jones et al. 2009
- Parameters:
- paramsstr | dict | None
The path to the parameter file for constructing the network. If None, parameters loaded from default.json Default: None
- add_drives_from_paramsbool
If True, add drives as defined in the params-dict. NB this is mainly for backward-compatibility with HNN GUI, and will be deprecated in a future release. Default: False
- legacy_modebool
Set to False by default. Enables matching HNN GUI output when drives are added suitably. Will be deprecated in a future release.
- mesh_shapetuple of int (default: (10, 10))
Defines the (n_x, n_y) shape of the grid of pyramidal cells.
- Returns:
- netInstance of Network object
Network object used to store
Notes
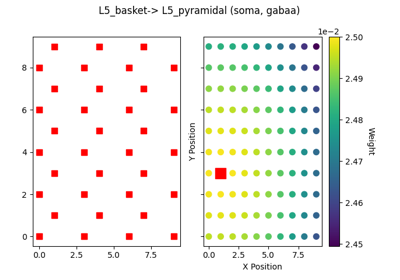
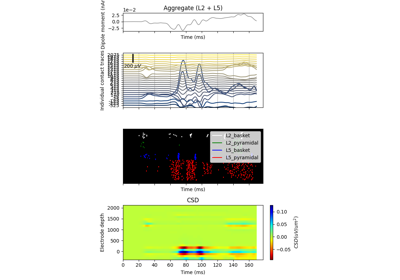
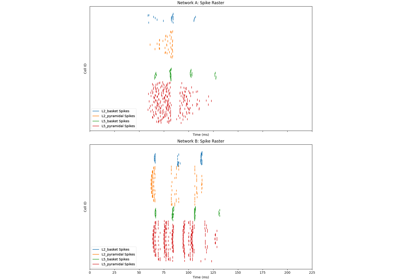
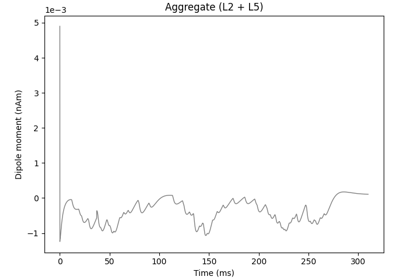
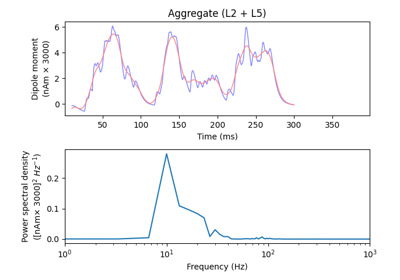
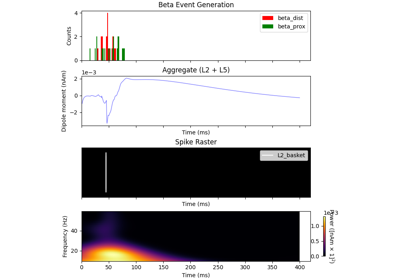
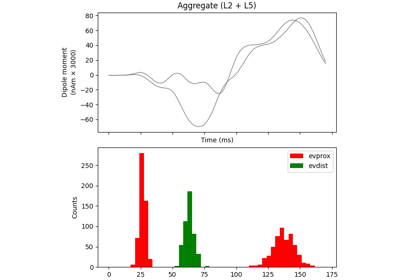
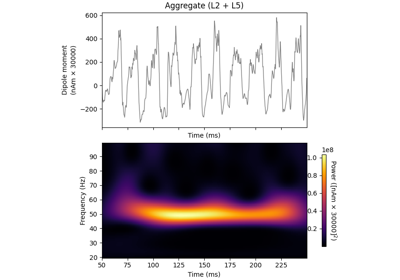
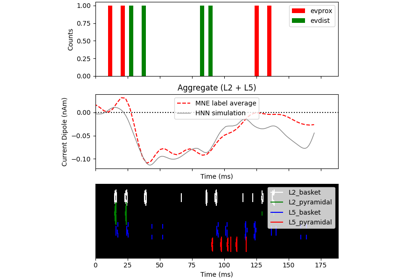
The network is composed of a square grid of pyramidal cells, arranged in two layers (L5 and L2). The default in-plane separation of the grid points is 1.0 um, and the layer separation 1307.4 um. These can be adjusted after the net is created using the set_cell_positions-method. An all-to-all connectivity pattern is applied between cells. Inhibitory basket cells are present at a 1:3-ratio.
This network was first described in Jones et al. 2009 [1] , and this code provides the implementation used in Neymotin et al. 2020 [2] .
References
[1]Jones, Stephanie R., et al. “Quantitative Analysis and Biophysically Realistic Neural Modeling of the MEG Mu Rhythm: Rhythmogenesis and Modulation of Sensory-Evoked Responses.” Journal of Neurophysiology 102, 3554–3572 (2009). https://doi.org/10.1152/jn.00535.2009
[2]Neymotin, Samuel A, et al. 2020. “Human Neocortical Neurosolver (HNN), a New Software Tool for Interpreting the Cellular and Network Origin of Human MEG/EEG Data.” eLife 9 (January):e51214. https://doi.org/10.7554/eLife.51214