Note
Click here to download the full example code or to run this example in your browser via Binder
06. Use MPI backend for parallelization¶
This example demonstrates how to use the MPI backend for simulating dipoles using HNN-core.
The MPI backend allows running the simulation in parallel across neurons in the network even with a single trial. For this, you will need the MPI related software installed. Note that if you want to simulate in parallel across trials, the Joblib backend allows this without the need to install and configure MPI.
# Authors: Mainak Jas <mainak.jas@telecom-paristech.fr>
# Blake Caldwell <blake_caldwell@brown.edu>
Let us import hnn_core
import os.path as op
import hnn_core
from hnn_core import simulate_dipole, read_params, Network
Then we setup the directories and Neuron
hnn_core_root = op.dirname(hnn_core.__file__)
Then we read the default parameters file
params_fname = op.join(hnn_core_root, 'param', 'default.json')
params = read_params(params_fname)
print(params)
Out:
{
"L2Basket_Gauss_A_weight": 0.0,
"L2Basket_Gauss_mu": 2000.0,
"L2Basket_Gauss_sigma": 3.6,
"L2Basket_Pois_A_weight_ampa": 0.0,
"L2Basket_Pois_A_weight_nmda": 0.0,
"L2Basket_Pois_lamtha": 0.0,
"L2Pyr_Gauss_A_weight": 0.0,
"L2Pyr_Gauss_mu": 2000.0,
"L2Pyr_Gauss_sigma": 3.6,
"L2Pyr_Pois_A_weight_ampa": 0.0,
"L2Pyr_Pois_A_weight_nmda": 0.0,
"L2Pyr_Pois_lamtha": 0.0,
"L2Pyr_ampa_e": 0.0,
"L2Pyr_ampa_tau1": 0.5,
"L2Pyr_ampa_tau2": 5.0,
"L2Pyr_apical1_L": 306.0,
"L2Pyr_apical1_diam": 4.08,
"L2Pyr_apicaloblique_L": 340.0,
"L2Pyr_apicaloblique_diam": 3.91,
"L2Pyr_apicaltrunk_L": 59.5,
"L2Pyr_apicaltrunk_diam": 4.25,
"L2Pyr_apicaltuft_L": 238.0,
"L2Pyr_apicaltuft_diam": 3.4,
"L2Pyr_basal1_L": 85.0,
"L2Pyr_basal1_diam": 4.25,
"L2Pyr_basal2_L": 255.0,
"L2Pyr_basal2_diam": 2.72,
"L2Pyr_basal3_L": 255.0,
"L2Pyr_basal3_diam": 2.72,
"L2Pyr_dend_Ra": 200.0,
"L2Pyr_dend_cm": 0.6195,
"L2Pyr_dend_el_hh2": -65.0,
"L2Pyr_dend_gbar_km": 250.0,
"L2Pyr_dend_gkbar_hh2": 0.01,
"L2Pyr_dend_gl_hh2": 4.26e-05,
"L2Pyr_dend_gnabar_hh2": 0.15,
"L2Pyr_gabaa_e": -80.0,
"L2Pyr_gabaa_tau1": 0.5,
"L2Pyr_gabaa_tau2": 5.0,
"L2Pyr_gabab_e": -80.0,
"L2Pyr_gabab_tau1": 1.0,
"L2Pyr_gabab_tau2": 20.0,
"L2Pyr_nmda_e": 0.0,
"L2Pyr_nmda_tau1": 1.0,
"L2Pyr_nmda_tau2": 20.0,
"L2Pyr_soma_L": 22.1,
"L2Pyr_soma_Ra": 200.0,
"L2Pyr_soma_cm": 0.6195,
"L2Pyr_soma_diam": 23.4,
"L2Pyr_soma_el_hh2": -65.0,
"L2Pyr_soma_gbar_km": 250.0,
"L2Pyr_soma_gkbar_hh2": 0.01,
"L2Pyr_soma_gl_hh2": 4.26e-05,
"L2Pyr_soma_gnabar_hh2": 0.18,
"L5Basket_Gauss_A_weight": 0.0,
"L5Basket_Gauss_mu": 2000.0,
"L5Basket_Gauss_sigma": 2.0,
"L5Basket_Pois_A_weight_ampa": 0.0,
"L5Basket_Pois_A_weight_nmda": 0.0,
"L5Basket_Pois_lamtha": 0.0,
"L5Pyr_Gauss_A_weight": 0.0,
"L5Pyr_Gauss_mu": 2000.0,
"L5Pyr_Gauss_sigma": 4.8,
"L5Pyr_Pois_A_weight_ampa": 0.0,
"L5Pyr_Pois_A_weight_nmda": 0.0,
"L5Pyr_Pois_lamtha": 0.0,
"L5Pyr_ampa_e": 0.0,
"L5Pyr_ampa_tau1": 0.5,
"L5Pyr_ampa_tau2": 5.0,
"L5Pyr_apical1_L": 680.0,
"L5Pyr_apical1_diam": 7.48,
"L5Pyr_apical2_L": 680.0,
"L5Pyr_apical2_diam": 4.93,
"L5Pyr_apicaloblique_L": 255.0,
"L5Pyr_apicaloblique_diam": 5.1,
"L5Pyr_apicaltrunk_L": 102.0,
"L5Pyr_apicaltrunk_diam": 10.2,
"L5Pyr_apicaltuft_L": 425.0,
"L5Pyr_apicaltuft_diam": 3.4,
"L5Pyr_basal1_L": 85.0,
"L5Pyr_basal1_diam": 6.8,
"L5Pyr_basal2_L": 255.0,
"L5Pyr_basal2_diam": 8.5,
"L5Pyr_basal3_L": 255.0,
"L5Pyr_basal3_diam": 8.5,
"L5Pyr_dend_Ra": 200.0,
"L5Pyr_dend_cm": 0.85,
"L5Pyr_dend_el_hh2": -71.0,
"L5Pyr_dend_gbar_ar": 1e-06,
"L5Pyr_dend_gbar_ca": 60.0,
"L5Pyr_dend_gbar_cat": 0.0002,
"L5Pyr_dend_gbar_kca": 0.0002,
"L5Pyr_dend_gbar_km": 200.0,
"L5Pyr_dend_gkbar_hh2": 0.01,
"L5Pyr_dend_gl_hh2": 4.26e-05,
"L5Pyr_dend_gnabar_hh2": 0.14,
"L5Pyr_dend_taur_cad": 20.0,
"L5Pyr_gabaa_e": -80.0,
"L5Pyr_gabaa_tau1": 0.5,
"L5Pyr_gabaa_tau2": 5.0,
"L5Pyr_gabab_e": -80.0,
"L5Pyr_gabab_tau1": 1.0,
"L5Pyr_gabab_tau2": 20.0,
"L5Pyr_nmda_e": 0.0,
"L5Pyr_nmda_tau1": 1.0,
"L5Pyr_nmda_tau2": 20.0,
"L5Pyr_soma_L": 39.0,
"L5Pyr_soma_Ra": 200.0,
"L5Pyr_soma_cm": 0.85,
"L5Pyr_soma_diam": 28.9,
"L5Pyr_soma_el_hh2": -65.0,
"L5Pyr_soma_gbar_ar": 1e-06,
"L5Pyr_soma_gbar_ca": 60.0,
"L5Pyr_soma_gbar_cat": 0.0002,
"L5Pyr_soma_gbar_kca": 0.0002,
"L5Pyr_soma_gbar_km": 200.0,
"L5Pyr_soma_gkbar_hh2": 0.01,
"L5Pyr_soma_gl_hh2": 4.26e-05,
"L5Pyr_soma_gnabar_hh2": 0.16,
"L5Pyr_soma_taur_cad": 20.0,
"N_pyr_x": 10,
"N_pyr_y": 10,
"N_trials": 1,
"T_pois": -1,
"celsius": 37.0,
"dipole_scalefctr": 3000,
"dipole_smooth_win": 30,
"distribution_dist": "normal",
"distribution_prox": "normal",
"dt": 0.025,
"dt_evprox0_evdist": -1,
"dt_evprox0_evprox1": -1,
"events_per_cycle_dist": 2,
"events_per_cycle_prox": 2,
"f_input_dist": 10.0,
"f_input_prox": 10.0,
"f_max_spec": 100,
"f_stdev_dist": 20.0,
"f_stdev_prox": 20.0,
"gbar_L2Basket_L2Basket": 0.02,
"gbar_L2Basket_L2Pyr_gabaa": 0.05,
"gbar_L2Basket_L2Pyr_gabab": 0.05,
"gbar_L2Basket_L5Pyr": 0.001,
"gbar_L2Pyr_L2Basket": 0.0005,
"gbar_L2Pyr_L2Pyr_ampa": 0.0005,
"gbar_L2Pyr_L2Pyr_nmda": 0.0005,
"gbar_L2Pyr_L5Basket": 0.00025,
"gbar_L2Pyr_L5Pyr": 0.00025,
"gbar_L5Basket_L5Basket": 0.02,
"gbar_L5Basket_L5Pyr_gabaa": 0.025,
"gbar_L5Basket_L5Pyr_gabab": 0.025,
"gbar_L5Pyr_L5Basket": 0.0005,
"gbar_L5Pyr_L5Pyr_ampa": 0.0005,
"gbar_L5Pyr_L5Pyr_nmda": 0.0005,
"gbar_evdist_1_L2Basket_ampa": 0.006562,
"gbar_evdist_1_L2Basket_nmda": 0.019482,
"gbar_evdist_1_L2Pyr_ampa": 7e-06,
"gbar_evdist_1_L2Pyr_nmda": 0.004317,
"gbar_evdist_1_L5Pyr_ampa": 0.1423,
"gbar_evdist_1_L5Pyr_nmda": 0.080074,
"gbar_evprox_1_L2Basket_ampa": 0.08831,
"gbar_evprox_1_L2Basket_nmda": 0.0,
"gbar_evprox_1_L2Pyr_ampa": 0.01525,
"gbar_evprox_1_L2Pyr_nmda": 0.0,
"gbar_evprox_1_L5Basket_ampa": 0.19934,
"gbar_evprox_1_L5Basket_nmda": 0.0,
"gbar_evprox_1_L5Pyr_ampa": 0.00865,
"gbar_evprox_1_L5Pyr_nmda": 0.0,
"gbar_evprox_2_L2Basket_ampa": 3e-06,
"gbar_evprox_2_L2Basket_nmda": 0.0,
"gbar_evprox_2_L2Pyr_ampa": 1.43884,
"gbar_evprox_2_L2Pyr_nmda": 0.0,
"gbar_evprox_2_L5Basket_ampa": 0.008958,
"gbar_evprox_2_L5Basket_nmda": 0.0,
"gbar_evprox_2_L5Pyr_ampa": 0.684013,
"gbar_evprox_2_L5Pyr_nmda": 0.0,
"inc_evinput": 0.0,
"input_dist_A_delay_L2": 5.0,
"input_dist_A_delay_L5": 5.0,
"input_dist_A_weight_L2Basket_ampa": 0.0,
"input_dist_A_weight_L2Basket_nmda": 0.0,
"input_dist_A_weight_L2Pyr_ampa": 0.0,
"input_dist_A_weight_L2Pyr_nmda": 0.0,
"input_dist_A_weight_L5Pyr_ampa": 0.0,
"input_dist_A_weight_L5Pyr_nmda": 0.0,
"input_prox_A_delay_L2": 0.1,
"input_prox_A_delay_L5": 1.0,
"input_prox_A_weight_L2Basket_ampa": 0.0,
"input_prox_A_weight_L2Basket_nmda": 0.0,
"input_prox_A_weight_L2Pyr_ampa": 0.0,
"input_prox_A_weight_L2Pyr_nmda": 0.0,
"input_prox_A_weight_L5Basket_ampa": 0.0,
"input_prox_A_weight_L5Basket_nmda": 0.0,
"input_prox_A_weight_L5Pyr_ampa": 0.0,
"input_prox_A_weight_L5Pyr_nmda": 0.0,
"numspikes_evdist_1": 1,
"numspikes_evprox_1": 1,
"numspikes_evprox_2": 1,
"prng_seedcore_evdist_1": 2,
"prng_seedcore_evprox_1": 2,
"prng_seedcore_evprox_2": 2,
"prng_seedcore_extgauss": 2,
"prng_seedcore_extpois": 2,
"prng_seedcore_input_dist": 2,
"prng_seedcore_input_prox": 2,
"record_isoma": 0,
"record_vsoma": 0,
"repeats_dist": 10,
"repeats_prox": 10,
"save_dpl": 0,
"save_figs": 0,
"save_spec_data": 0,
"sigma_t_evdist_1": 3.85,
"sigma_t_evprox_1": 2.47,
"sigma_t_evprox_2": 8.33,
"sim_prefix": "default",
"spec_cmap": "jet",
"sync_evinput": false,
"t0_input_dist": 1000,
"t0_input_prox": 1000.0,
"t0_input_stdev_dist": 0.0,
"t0_input_stdev_prox": 0.0,
"t0_pois": 0.0,
"t_evdist_1": 63.53,
"t_evprox_1": 26.61,
"t_evprox_2": 137.12,
"threshold": 0.0,
"tstop": 170,
"tstop_input_dist": 1001,
"tstop_input_prox": 1001
}
Update a few of the default parameters related to visualisation
params.update({
'tstop': 310.0,
})
Following the alpha example, we add a ~10 Hz “bursty” drive starting at 50 ms and continuing to the end of the simulation. Each burst consists of a pair (2) of spikes, spaced 10 ms apart. The occurrence of each burst is jittered by a random, normally distributed amount (20 ms standard deviation). We repeat the burst train 10 times, each time with unique randomization.
net = Network(params)
weights_ampa = {'L2_pyramidal': 5.4e-5, 'L5_pyramidal': 5.4e-5}
net.add_bursty_drive(
'bursty', tstart=50., burst_rate=10, burst_std=20., numspikes=2,
spike_isi=10, repeats=10, location='distal', weights_ampa=weights_ampa,
seedcore=4)
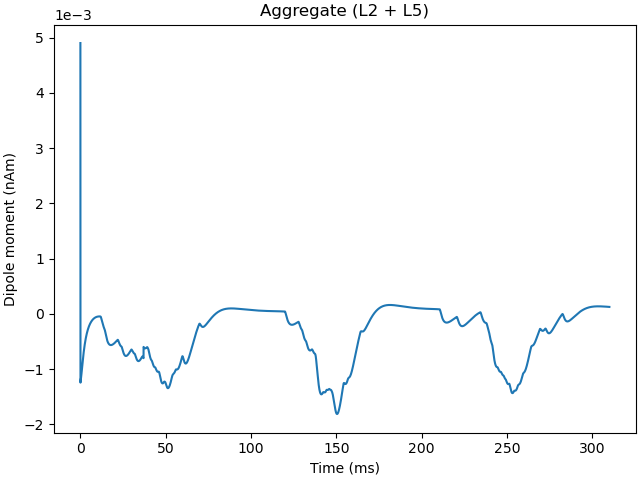
Finally, to simulate we use the
MPIBackend class. This will
start the simulation across the number of processors (cores) specified by
n_procs using MPI. The 'mpiexec' launcher is used from
openmpi, which must be installed on the system
Out:
MPI will run over 2 processes
Running 1 trials...
numprocs=2
[1d68482510bd:01667] Read -1, expected 58804, errno = 1
Loading custom mechanism files from /home/circleci/miniconda/envs/testenv/lib/python3.6/site-packages/hnn_core-0.1-py3.6.egg/hnn_core/mod/x86_64/.libs/libnrnmech.so
Building the NEURON model
Loading custom mechanism files from /home/circleci/miniconda/envs/testenv/lib/python3.6/site-packages/hnn_core-0.1-py3.6.egg/hnn_core/mod/x86_64/.libs/libnrnmech.so
[Done]
running trial 1 on 2 cores
Simulation time: 0.03 ms...
Simulation time: 10.0 ms...
Simulation time: 20.0 ms...
Simulation time: 30.0 ms...
Simulation time: 40.0 ms...
Simulation time: 50.0 ms...
Simulation time: 60.0 ms...
Simulation time: 70.0 ms...
Simulation time: 80.0 ms...
Simulation time: 90.0 ms...
Simulation time: 100.0 ms...
Simulation time: 110.0 ms...
Simulation time: 120.0 ms...
Simulation time: 130.0 ms...
Simulation time: 140.0 ms...
Simulation time: 150.0 ms...
Simulation time: 160.0 ms...
Simulation time: 170.0 ms...
Simulation time: 180.0 ms...
Simulation time: 190.0 ms...
Simulation time: 200.0 ms...
Simulation time: 210.0 ms...
Simulation time: 220.0 ms...
Simulation time: 230.0 ms...
Simulation time: 240.0 ms...
Simulation time: 250.0 ms...
Simulation time: 260.0 ms...
Simulation time: 270.0 ms...
Simulation time: 280.0 ms...
Simulation time: 290.0 ms...
Simulation time: 300.0 ms...
[1d68482510bd:01667] Read -1, expected 49608, errno = 1
[1d68482510bd:01666] Read -1, expected 49600, errno = 1
[1d68482510bd:01667] Read -1, expected 49600, errno = 1
[1d68482510bd:01666] Read -1, expected 49608, errno = 1
[1d68482510bd:01667] Read -1, expected 49608, errno = 1
[1d68482510bd:01666] Read -1, expected 49600, errno = 1
[1d68482510bd:01667] Read -1, expected 49600, errno = 1
[1d68482510bd:01666] Read -1, expected 49608, errno = 1
[1d68482510bd:01666] Read -1, expected 5616, errno = 1
<Figure size 640x480 with 1 Axes>
Total running time of the script: ( 0 minutes 35.615 seconds)