Note
Go to the end to download the full example code or to run this example in your browser via Binder.
09. Replaying Spike Data as Input#
Welcome to this hands-on tutorial on simulating communication between two neocortical networks using HNN-Core! This notebook guides you through creating a source network (Network A), extracting its spike data, feeding it into a target network (Network B) using the add_spike_train_drive method, simulating both, and visualizing the results. Let’s dive in!
# Authors: Maira Usman <maira.usman5703@gmail.com>
Step 1: Setting Up the Source Network (Network A)#
Our journey begins with Network A, the source of neural activity. We’ll use the jones_2009_model to create a realistic neocortical network and simulate it with an evoked drive to generate spike data. This step mimics how a network might produce activity in a real brain region, which we’ll later transmit to another network.
The simulation runs for 170 ms, and we’ll save the spike data to a temporary file for efficient handling.
import numpy as np
import matplotlib.pyplot as plt
from hnn_core import jones_2009_model, simulate_dipole, read_spikes
import os.path as op
import tempfile
# Create and simulate Network A
print("Creating and simulating Network A...")
net_A = jones_2009_model()
net_A._params.update({'tstop': 170.0})
# Add a drive to generate activity
print("Adding drive to Network A...")
drive_params = {
'name': 'evdist1', 'mu': 63.5, 'sigma': 3.8, 'numspikes': 1,
'location': 'distal', 'event_seed': 274
}
weights = {
'ampa': {'L2_basket': 0.006, 'L2_pyramidal': 0.0005, 'L5_pyramidal': 0.14},
'nmda': {'L2_basket': 0.019, 'L2_pyramidal': 0.004, 'L5_pyramidal': 0.08}
}
delays = {'L2_basket': 0.1, 'L2_pyramidal': 0.1, 'L5_pyramidal': 0.1}
net_A.add_evoked_drive(
**drive_params, weights_ampa=weights['ampa'], weights_nmda=weights['nmda'],
synaptic_delays=delays
)
# Simulate and save spike data
print("Simulating Network A and saving spikes...")
dpls_A = simulate_dipole(net_A, tstop=170.0, n_trials=1)
with tempfile.TemporaryDirectory() as tmp_dir:
spike_file = op.join(tmp_dir, 'spk_%d.txt')
net_A.cell_response.write(spike_file)
cell_response = read_spikes(op.join(tmp_dir, 'spk_*.txt'))
print(f"Extracted {sum(len(t) for t in cell_response.spike_times)} spikes from Network A")
Creating and simulating Network A...
Adding drive to Network A...
Simulating Network A and saving spikes...
Joblib will run 1 trial(s) in parallel by distributing trials over 1 jobs.
Building the NEURON model
[Done]
Trial 1: 0.03 ms...
Trial 1: 10.0 ms...
Trial 1: 20.0 ms...
Trial 1: 30.0 ms...
Trial 1: 40.0 ms...
Trial 1: 50.0 ms...
Trial 1: 60.0 ms...
Trial 1: 70.0 ms...
Trial 1: 80.0 ms...
Trial 1: 90.0 ms...
Trial 1: 100.0 ms...
Trial 1: 110.0 ms...
Trial 1: 120.0 ms...
Trial 1: 130.0 ms...
Trial 1: 140.0 ms...
Trial 1: 150.0 ms...
Trial 1: 160.0 ms...
Writing file /tmp/tmpsm208jmo/spk_0.txt
/home/circleci/project/examples/howto/plot_replaying_spike_data_as_input.py:59: DeprecationWarning: Reading cell response from txt files is deprecated and will be removed in future versions. Please load cell response along with simulated network
cell_response = read_spikes(op.join(tmp_dir, 'spk_*.txt'))
Extracted 616 spikes from Network A
Step 2: Configuring the Target Network (Network B)#
Now, let’s set up Network B, the target network that will receive spike data from Network A. We’ll extract spikes from the first trial of Network A, filter for pyramidal cells, and format them into a dictionary compatible with add_spike_train_drive. This process is akin to distributing computed data across nodes in an MPI setup, where each node (here, Network B) processes a subset of the input.
We’ll also define connection properties to map Network A’s pyramidal cells to Network B’s layers, ensuring a realistic interaction.
print("Creating and configuring Network B...")
net_B = jones_2009_model()
net_B._params.update({'tstop': 225.0})
# Extract spike data from first trial and filter for pyramidal cells
trial_idx = 0
spike_times = cell_response.spike_times[trial_idx]
spike_gids = cell_response.spike_gids[trial_idx]
spike_types = cell_response.spike_types[trial_idx]
pyramidal_mask = np.array([t in ['L2_pyramidal', 'L5_pyramidal'] for t in spike_types])
filtered_times = np.array(spike_times)[pyramidal_mask]
filtered_gids = np.array(spike_gids)[pyramidal_mask]
filtered_types = np.array(spike_types)[pyramidal_mask]
spike_data = {f"NetA_{t}_GID{g}": [] for t, g in zip(filtered_types, filtered_gids)}
for t, g, time in zip(filtered_types, filtered_gids, filtered_times):
src_id = f"NetA_{t}_GID{g}"
spike_data[src_id].append(time)
print(f"Number of valid pyramidal cells with spikes to be used for input: {len(spike_data)}")
# Define target configuration
conn_properties = {
'L5_pyramidal': {'location': 'distal', 'weights_ampa': 0.005, 'synaptic_delays': 1.5},
'L2_pyramidal': {'location': 'proximal', 'weights_ampa': 0.003, 'synaptic_delays': 1.0}
}
target_config = {}
source_ids = list(spike_data.keys())
for i, src_id in enumerate(source_ids):
target_type = 'L5_pyramidal' if i % 2 == 0 else 'L2_pyramidal'
props = conn_properties[target_type]
target_config[src_id] = {
'target_cell_types': [target_type],
'location': props['location'],
'weights_ampa': {target_type: props['weights_ampa']},
'synaptic_delays': {target_type: props['synaptic_delays']},
'probability': 0.7
}
all_weights_ampa = {k: cfg['weights_ampa'][k] for cfg in target_config.values() for k in cfg['weights_ampa']}
# Add spike train drive to Network B
net_B.add_spike_train_drive(
name='drive_from_NetA',
spike_data=spike_data,
location='distal',
weights_ampa=all_weights_ampa,
weights_nmda=None,
synaptic_delays=0.1,
conn_seed=42
)
Creating and configuring Network B...
Number of valid pyramidal cells with spikes to be used for input: 139
/home/circleci/miniconda/envs/testenv/lib/python3.11/site-packages/hnn_core/network.py:1200: UserWarning: Spike train drives can only target sections defined in `Cell.sect_loc` when `legacy_mode=False`.
warnings.warn(
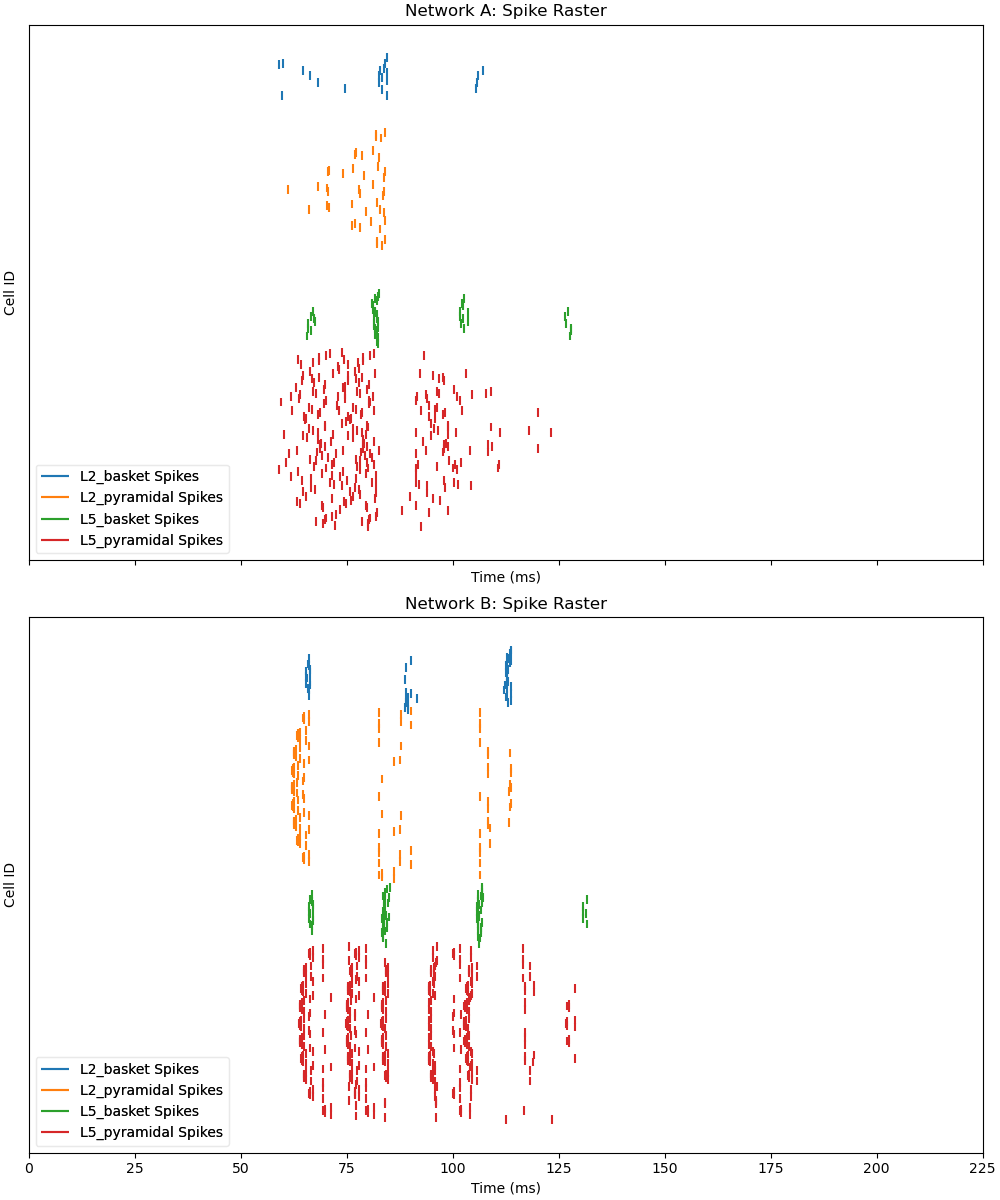
Step 3: Simulating and Visualizing the Networks#
With Network B configured, we’ll simulate it for 225 ms to observe the effect of Network A’s spikes. Visualization is key in neuroscience, so we’ll create raster plots for both networks. This step lets you see how the input from Network A influences Network B’s activity.
print("Simulating Network B...")
dpls_b = simulate_dipole(net_B, tstop=225.0, n_trials=1)
# Visualize results
fig, axes = plt.subplots(2, 1, figsize=(10, 12), sharex=True, constrained_layout=True)
# 1. Network A: Spike raster
net_A.cell_response.plot_spikes_raster(ax=axes[0], show=False)
axes[0].set_title('Network A: Spike Raster')
axes[0].set_ylabel('Cell ID')
# 2. Network B: Spike raster
net_B.cell_response.plot_spikes_raster(ax=axes[1], show=False)
axes[1].set_title('Network B: Spike Raster')
axes[1].set_ylabel('Cell ID')
axes[1].set_xlabel('Time (ms)')
plt.show()
Simulating Network B...
Joblib will run 1 trial(s) in parallel by distributing trials over 1 jobs.
Building the NEURON model
[Done]
Trial 1: 0.03 ms...
Trial 1: 10.0 ms...
Trial 1: 20.0 ms...
Trial 1: 30.0 ms...
Trial 1: 40.0 ms...
Trial 1: 50.0 ms...
Trial 1: 60.0 ms...
Trial 1: 70.0 ms...
Trial 1: 80.0 ms...
Trial 1: 90.0 ms...
Trial 1: 100.0 ms...
Trial 1: 110.0 ms...
Trial 1: 120.0 ms...
Trial 1: 130.0 ms...
Trial 1: 140.0 ms...
Trial 1: 150.0 ms...
Trial 1: 160.0 ms...
Trial 1: 170.0 ms...
Trial 1: 180.0 ms...
Trial 1: 190.0 ms...
Trial 1: 200.0 ms...
Trial 1: 210.0 ms...
Trial 1: 220.0 ms...
Now that the simulation is finished, let’s verify the drive setup and ensure the connection was established correctly.
print("\nVerifying configuration:")
drive_name = 'drive_from_NetA'
if drive_name in net_B.external_drives:
drive = net_B.external_drives[drive_name]
print(f" Drive '{drive_name}' created with {drive['n_drive_cells']} cells")
if drive_name in net_B.gid_ranges:
print(f" GIDs assigned: {net_B.gid_ranges[drive_name]}")
drive_connections = sum(1 for conn in net_B.connectivity if conn['src_type'] == drive_name)
print(f" Number of connections from drive: {drive_connections}")
else:
print(f" ERROR: Drive '{drive_name}' not found")
Verifying configuration:
Drive 'drive_from_NetA' created with 139 cells
GIDs assigned: range(270, 409)
Number of connections from drive: 2
Conclusion#
You’ve now successfully replayed spikes from one network into another network! The raster plots show how Network A’s activity influences Network B, while the verification confirms the drive setup. This approach is scalable—similar to MPI’s distributed computing—allowing you to experiment with multiple networks or adjust parameters like weights and delays to explore different dynamics. Try modifying the weights_ampa or synaptic_delays values in Step 2 to see how they affect the results!
Total running time of the script: (1 minutes 11.969 seconds)
