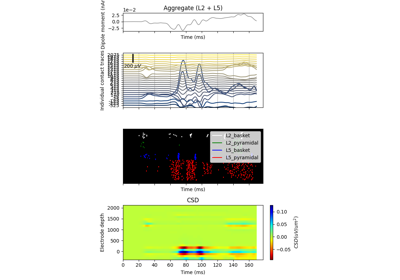
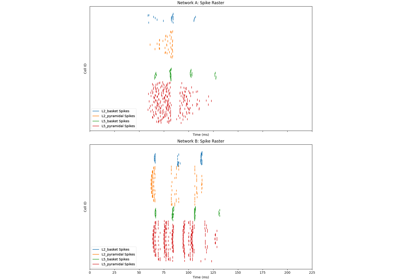
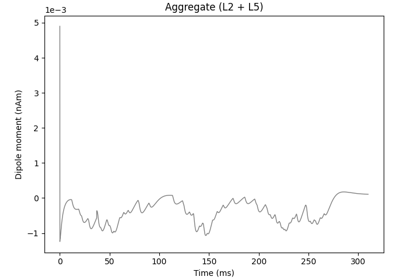
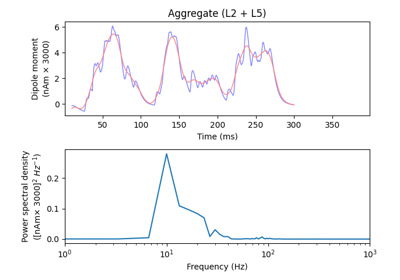
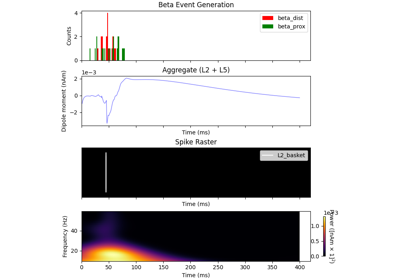
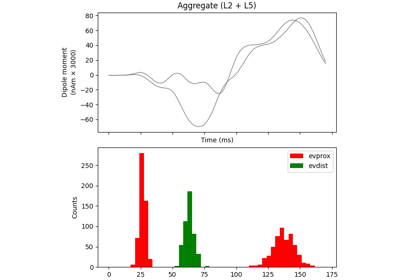
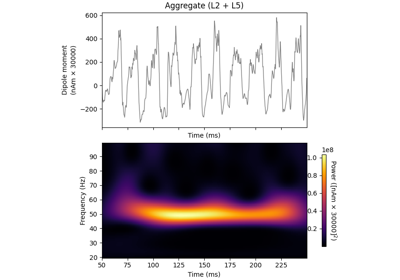
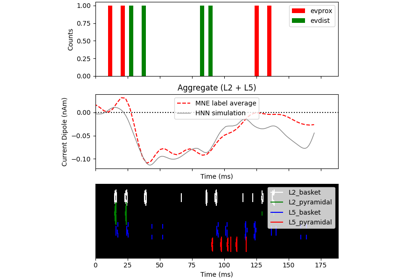
hnn_core.Network#
- class hnn_core.Network(params, add_drives_from_params=False, legacy_mode=False, mesh_shape=(10, 10), pos_dict=None, cell_types=None)[source]#
The Network class.
- Parameters:
- paramsdict
The parameters to use for constructing the network.
- add_drives_from_paramsbool, default=False
If True, add drives as defined in the params-dict. NB this is mainly for backward-compatibility with HNN GUI, and will be deprecated in a future release.
- legacy_modebool, default=False
Set to True by default to enable matching HNN GUI output when drives are added suitably. Will be deprecated in a future release.
- mesh_shapetuple of int (default: (10, 10))
Defines the (n_x, n_y) shape of the grid of pyramidal cells.
- pos_dictdict of list of tuple (x, y, z), optional
Dictionary containing the coordinate positions of all cells. Keys are ‘L2_pyramidal’, ‘L5_pyramidal’, ‘L2_basket’, ‘L5_basket’, or any external drive name.
- cell_typesdict of dict of (Cell | dict), optional
Dictionary containing names of real cell types in the network (e.g. ‘L2_basket’) as keys and child-dictionaries describing the cell type. The child-dictionary contains two keys: “cell_object” and “cell_metadata”. The value of “cell_object” is the corresponding Cell instance of the cell type being described, and this instance is used as a template for the other cells of its type in the population. The value of “cell_metadata” is a dictionary containing several key-values pairs that describe different aspects of the cell type, described below:
“morpho_type” : either “basket” or “pyramidal”
“electro_type” : either “inhibitory” or “excitatory”
“layer” : either “2” or “5”
“measure_dipole” : either True or False
“reference”: optional
- Attributes:
- cell_typesdict of dict of (Cell | dict)
Dictionary containing names of real cell types in the network (e.g. ‘L2_basket’) as keys and child-dictionaries describing the cell type. The child-dictionary contains two keys: “cell_object” and “cell_metadata”. The value of “cell_object” is the corresponding Cell instance of the cell type being described, and this instance is used as a template for the other cells of its type in the population. The value of “cell_metadata” is a dictionary containing several key-values pairs that describe different aspects of the cell type, described below:
“morpho_type” : either “basket” or “pyramidal”
“electro_type” : either “inhibitory” or “excitatory”
“layer” : either “2” or “5”
“measure_dipole” : either True or False
“reference”: optional
- gid_rangesdict
A dictionary of unique identifiers of each real and artificial cell in the network. Every cell type is represented by a key read from cell_types, followed by keys read from external_drives. The value of each key is a range of ints, one for each cell in given category. Examples: ‘L2_basket’: range(0, 270), ‘evdist1’: range(272, 542), etc
- pos_dictdict of list of tuple (x, y, z)
Dictionary containing the coordinate positions of all cells. Keys are ‘L2_pyramidal’, ‘L5_pyramidal’, ‘L2_basket’, ‘L5_basket’, or any external drive name.
- cell_responseCellResponse
An instance of the CellResponse object.
- external_drivesdict (keys: drive names) of dict (keys: parameters)
The external driving inputs to the network. Drives are added by defining their spike-time dynamics, and their connectivity to the real cells of the network. Event times are instantiated before simulation, and are stored under the
'events'-key (list of list; first index for trials, second for event time lists for each drive cell).- external_biasesdict of dict (bias parameters for each cell type)
The parameters of bias inputs to cell somata, e.g., tonic current clamp
- connectivitylist of dict
List of dictionaries specifying each cell-cell and drive-cell connection
- rec_arraysdict
Stores electrode position information and voltages recorded by them for extracellular potential measurements. Multiple electrode arrays may be defined as unique keys. The values of the dictionary are instances of
hnn_core.extracellular.ExtracellularArray.- thresholdfloat
Firing threshold of all cells.
- delayfloat
Synaptic delay in ms.
Methods
add_bursty_drive(name, *[, tstart, ...])Add a bursty (rhythmic) external drive to all cells of the network
add_connection(src_gids, target_gids, loc, ...)Appends connections to connectivity list
add_electrode_array(name, electrode_pos, *)Specify coordinates of electrode array for extracellular recording.
add_evoked_drive(name, *, mu, sigma, ...[, ...])Add an 'evoked' external drive to the network
add_poisson_drive(name, *[, tstart, tstop, ...])Add a Poisson-distributed external drive to the network
add_spike_train_drive(name, *, spike_data, ...)Add an external drive from explicitly defined spike trains.
add_tonic_bias(*[, cell_type, section, ...])Attaches parameters of tonic bias input for given cell types
Remove all connections defined in Network.connectivity
Remove all drives defined in Network.connectivity
copy()Return a copy of the Network instance
filter_cell_types(**metadata_filters)Filter cell types based on cell_metadata criteria
Retrieve gain values for different celltype connections in the Network.
gid_to_type(gid)Reverse lookup of gid to type.
plot_cells([ax, show])Plot the cells using Network.pos_dict.
set_cell_positions(*[, inplane_distance, ...])Set relative positions of cells arranged in a square grid
set_global_synaptic_gains([e_e, e_i, i_e, ...])Change the synaptic gains of the celltypes in the Network.
write_configuration(fname[, overwrite])Writes network configuration to a json file.
to_dict
Notes
net = jones_2009_model(params)is the recommended path for creating a network. Instantiating the network asnet = Network(params)will produce a network with no cell-to-cell connections. As such, connectivity information contained inparamswill be ignored.- add_bursty_drive(name, *, tstart=0, tstart_std=0, tstop=None, location, burst_rate, burst_std=0, numspikes=2, spike_isi=10, n_drive_cells=1, cell_specific=False, weights_ampa=None, weights_nmda=None, synaptic_delays=0.1, space_constant=100.0, probability=1.0, event_seed=2, conn_seed=3)[source]#
Add a bursty (rhythmic) external drive to all cells of the network
- Parameters:
- namestr
Unique name for the drive
- tstartfloat
Start time of the burst trains (default: 0)
- tstart_stdfloat
If greater than 0, randomize start time with standard deviation tstart_std (unit: ms). Effectively jitters start time across multiple trials.
- tstopfloat
End time of burst trains (defaults to None: tstop is set to the end of the simulation)
- locationstr
Target location of synapses. Must be an element of Cell.sect_loc such as ‘proximal’ or ‘distal’, which defines a group of sections, or an existing section such as ‘soma’ or ‘apical_tuft’ (defined in Cell.sections for all targeted cells). The parameter legacy_mode of the
Networkmust be set to False to target specific sections.- burst_ratefloat
The mean rate at which cyclic bursts occur (unit: Hz)
- burst_stdfloat
The standard deviation of the burst occurrence on each cycle (unit: ms). Default: 0 ms
- numspikesint
The number of spikes in a burst. This is the spikes/burst parameter in the GUI. Default: 2 (doublet)
- spike_isifloat
Time between spike events within a cycle (ISI). Default: 10 ms
- n_drive_cellsint | ‘n_cells’
The number of drive cells that contribute an independently sampled burst at each cycle. If n_drive_cells=’n_cells’ and cell_specific=True, a drive cell gets assigned to each available simulated cell in the network with 1-to-1 connectivity. Otherwise (default: 1), drive cells are assigned with all-to-all connectivity and provide synchronous input to cells in the network.
- cell_specificbool
Whether each artificial drive cell has 1-to-1 (True) or all-to-all (False, default) connection parameters. Note that 1-to-1 connectivity requires that n_drive_cells=’n_cells’, where ‘n_cells’ denotes the number of all available cells that this drive can target in the network.
- weights_ampadict or None
Synaptic weights (in uS) of AMPA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- weights_nmdadict or None
Synaptic weights (in uS) of NMDA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- synaptic_delaysdict or float
Synaptic delay (in ms) at the column origin, dispersed laterally as a function of the space_constant. If float, applies to all target cell types. Use dict to create delay->cell mapping.
- space_constantfloat
Describes lateral dispersion (from the column origin) of synaptic weights and delays within the simulated column. The constant is measured in the units of
inplane_distanceofNetwork. For example, forspace_constant=3, the weights and delays are modulated by the factorexp(-(x / (3 * inplane_distance)) ** 2), wherexis the physical distance (in um) between the connected cells in the xy plane.- probabilitydict or float (default: 1.0)
Probability of connection between any src-target pair. Use dict to create probability->cell mapping. If float, applies to all target cell types.
- event_seedint
Optional initial seed for random number generator (default: 2). Used to generate event times for drive cells.
- conn_seedint
Optional initial seed for random number generator (default: 3). Used to randomly remove connections when probability < 1.0.
- add_connection(src_gids, target_gids, loc, receptor, weight, delay, lamtha, threshold=None, gain=1.0, allow_autapses=True, probability=1.0, conn_seed=None)[source]#
Appends connections to connectivity list
- Parameters:
- src_gidsstr | int | range | list of int
Identifier for source cells. Passing str arguments (‘evdist1’, ‘L2_pyramidal’, ‘L2_basket’, ‘L5_pyramidal’, ‘L5_basket’, etc.) is equivalent to passing a list of gids for the relevant cell type. source - target connections are made in an all-to-all pattern.
- target_gidsstr | int | range | list of int
Identifier for targets of source cells. Passing str arguments (‘L2_pyramidal’, ‘L2_basket’, ‘L5_pyramidal’, ‘L5_basket’) is equivalent to passing a list of gids for the relevant cell type. source - target connections are made in an all-to-all pattern.
- locstr
Target location of synapses. Must be an element of Cell.sect_loc such as ‘proximal’ or ‘distal’, which defines a group of sections, or an existing section such as ‘soma’ or ‘apical_tuft’ (defined in Cell.sections for all targeted cells). The parameter legacy_mode of the
Networkmust be set to False to target specific sections.- receptorstr
Synaptic receptor of connection. Must be one of: ‘ampa’, ‘nmda’, ‘gabaa’, or ‘gabab’.
- weightfloat
Synaptic weight on target cell.
- delayfloat
Synaptic delay in ms.
- lamthafloat
Space constant.
- thresholdfloat, default=None
Firing threshold of cells for connection. If None (the default), inherit the threshold from the Network object.
- gainfloat, default=1.0
Multiplicative factor for synaptic weight.
- allow_autapsesbool, default=True
If True, allow connecting neuron to itself.
- probabilityfloat, default=1.0
Probability of connection between any src-target pair. Defaults to 1.0 producing an all-to-all pattern.
- conn_seedint, default=None
Optional initial seed for random number generator (default: None). Used to randomly remove connections when probability < 1.0.
Notes
Connections are stored in
net.connectivity[idx]['gid_pairs'], a dictionary indexed by src gids with the format: {src_gid: [target_gids, …], …} where each src_gid indexes a list of all its targets.
- add_electrode_array(name, electrode_pos, *, conductivity=0.3, method='psa', min_distance=0.5)[source]#
Specify coordinates of electrode array for extracellular recording.
- Parameters:
- namestr
Unique name of the array.
- electrode_postuple | list of tuple
Coordinates specifying the position for extracellular electrodes in the form of (x, y, z) (in um).
- conductivityfloat
Extracellular conductivity, in S/m, of the assumed infinite, homogeneous volume conductor that the cell and electrode are in.
- methodstr
Approximation to use.
'psa'(point source approximation) treats each segment junction as a point extracellular current source.'lsa'(line source approximation) treats each segment as a line source of current, which extends from the previous to the next segment center point: /—x—/, where x is the current segment flanked by /.- min_distancefloat (default: 0.5; unit: um)
To avoid numerical errors in calculating potentials, apply a minimum distance limit between the electrode contacts and the active neuronal membrane elements that act as sources of current. The default value of 0.5 um corresponds to 1 um diameter dendrites.
- add_evoked_drive(name, *, mu, sigma, numspikes, location, n_drive_cells='n_cells', cell_specific=True, weights_ampa=None, weights_nmda=None, space_constant=3.0, synaptic_delays=0.1, probability=1.0, event_seed=2, conn_seed=3)[source]#
Add an ‘evoked’ external drive to the network
- Parameters:
- namestr
Unique name for the drive
- mufloat
Mean of Gaussian event time distribution
- sigmafloat
Standard deviation of event time distribution
- numspikesint
Number of spikes at each target cell
- locationstr
Target location of synapses. Must be an element of Cell.sect_loc such as ‘proximal’ or ‘distal’, which defines a group of sections, or an existing section such as ‘soma’ or ‘apical_tuft’ (defined in Cell.sections for all targeted cells). The parameter legacy_mode of the
Networkmust be set to False to target specific sections.- n_drive_cellsint | ‘n_cells’
The number of drive cells that each contribute an independently sampled synaptic spike to the network according to the Gaussian time distribution (mu, sigma). If n_drive_cells=’n_cells’ (default) and cell_specific=True, a drive cell gets assigned to each available simulated cell in the network with 1-to-1 connectivity. Otherwise, drive cells are assigned with all-to-all connectivity. If you wish to synchronize the timing of this evoked drive across the network in a given trial with one spike, set n_drive_cells=1 and cell_specific=False.
- cell_specificbool
Whether each artificial drive cell has 1-to-1 (True, default) or all-to-all (False) connection parameters. Note that 1-to-1 connectivity requires that n_drive_cells=’n_cells’, where ‘n_cells’ denotes the number of all available cells that this drive can target in the network.
- weights_ampadict or None
Synaptic weights (in uS) of AMPA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- weights_nmdadict or None
Synaptic weights (in uS) of NMDA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- synaptic_delaysdict or float
Synaptic delay (in ms) at the column origin, dispersed laterally as a function of the space_constant. If float, applies to all target cell types. Use dict to create delay->cell mapping.
- space_constantfloat
Describes lateral dispersion (from the column origin) of synaptic weights and delays within the simulated column. The constant is measured in the units of
inplane_distanceofNetwork. For example, forspace_constant=3, the weights are modulated by the factorexp(-(x / (3 * inplane_distance)) ** 2), where x is the physical distance (in um) between the connected cells in the xy plane (delays are modulated by the inverse of this factor).- probabilitydict or float (default: 1.0)
Probability of connection between any src-target pair. Use dict to create probability->cell mapping. If float, applies to all target cell types
- event_seedint
Optional initial seed for random number generator (default: 2). Used to generate event times for drive cells. Not fixed across trials (see Notes)
- conn_seedint
Optional initial seed for random number generator (default: 3). Used to randomly remove connections when probability < 1.0. Fixed across trials (see Notes)
Notes
Random seeding behavior across trials is different for event_seed and conn_seed (n_trials > 1 in simulate_dipole(…, n_trials):
- event_seed
Across trials, the random seed is incremented such that the exact spike times are different
- conn_seed
The random seed does not change across trials. This means for probability < 1.0, the random subset of gids targeted is the same.
- add_poisson_drive(name, *, tstart=0, tstop=None, rate_constant, location, n_drive_cells='n_cells', cell_specific=True, weights_ampa=None, weights_nmda=None, space_constant=100.0, synaptic_delays=0.1, probability=1.0, event_seed=2, conn_seed=3)[source]#
Add a Poisson-distributed external drive to the network
- Parameters:
- namestr
Unique name for the drive
- tstartfloat
Start time of Poisson-distributed spike train (default: 0)
- tstopfloat
End time of the spike train (defaults to None: tstop is set to the end of the simulation)
- rate_constantfloat or dict of floats
Rate constant (lambda > 0) of the renewal-process generating the samples. If a float is provided, the same rate constant is applied to each target cell type. Cell type-specific values may be provided as a dictionary, in which a key must be present for each cell type with non-zero AMPA or NMDA weights.
- locationstr
Target location of synapses. Must be an element of Cell.sect_loc such as ‘proximal’ or ‘distal’, which defines a group of sections, or an existing section such as ‘soma’ or ‘apical_tuft’ (defined in Cell.sections for all targeted cells). The parameter legacy_mode of the
Networkmust be set to False to target specific sections.- n_drive_cellsint | ‘n_cells’
The number of drive cells that each contribute an independently sampled synaptic spike to the network according to a Poisson process. If n_drive_cells=’n_cells’ (default) and cell_specific=True, a drive cell gets assigned to each available simulated cell in the network with 1-to-1 connectivity. Otherwise, drive cells are assigned with all-to-all connectivity. If you wish to synchronize the timing of Poisson drive across the network in a given trial, set n_drive_cells=1 and cell_specific=False.
- cell_specificbool
Whether each artificial drive cell has 1-to-1 (True, default) or all-to-all (False) connection parameters. Note that 1-to-1 connectivity requires that n_drive_cells=’n_cells’, where ‘n_cells’ denotes the number of all available cells that this drive can target in the network.
- weights_ampadict or None
Synaptic weights (in uS) of AMPA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- weights_nmdadict or None
Synaptic weights (in uS) of NMDA receptors on each targeted cell type (dict keys). Cell types omitted from the dict are set to zero.
- synaptic_delaysdict or float
Synaptic delay (in ms) at the column origin, dispersed laterally as a function of the space_constant. If float, applies to all target cell types. Use dict to create delay->cell mapping.
- space_constantfloat
Describes lateral dispersion (from the column origin) of synaptic weights and delays within the simulated column. The constant is measured in the units of
inplane_distanceofNetwork. For example, forspace_constant=3, the weights and delays are modulated by the factorexp(-(x / (3 * inplane_distance)) ** 2), wherexis the physical distance (in um) between the connected cells in the xy plane.- probabilitydict or float (default: 1.0)
Probability of connection between any src-target pair. Use dict to create probability->cell mapping. If float, applies to all target cell types.
- event_seedint
Optional initial seed for random number generator (default: 2). Used to generate event times for drive cells.
- conn_seedint
Optional initial seed for random number generator (default: 3). Used to randomly remove connections when probability < 1.0.
- add_spike_train_drive(name, *, spike_data, location, weights_ampa=None, weights_nmda=None, synaptic_delays=0.1, space_constant=3.0, probability=1.0, conn_seed=None)[source]#
Add an external drive from explicitly defined spike trains.
This method enables the target network to receive spike trains from a source network (e.g., another HNN simulation) or external data, driving activity in the target network’s cells.
- Parameters:
- namestr
Unique name for the drive (e.g., ‘drive_from_NetA’).
- spike_datadict or list of tuple
Spike train data from the source network (or external source) in one of three formats:
Format 1 (dictionary): Keys are unique identifiers (str) for source
cells and values are lists of spike times in milliseconds. The keys can be any string that helps identify the source. Example:
` {"NetA_L2_pyramidal_GID0": [10.2, 25.3], "NetA_L5_pyramidal_GID1": [15.1, 30.4]} `Format 2 (tuples): A list of (time, gid) tuples, where each tuple
contains a spike time (float, in ms) and a GID (int). The GIDs can be any integers that uniquely identify different source cells. Example:
` [(10.2, 0), (25.3, 1), (15.1, 0), (30.4, 1)] `Format 3: String path (or glob pattern) to spike files that can be loaded
with
read_spikes(). Example: “path/to/spk_*.txt”Note: The GIDs in both formats refer to the source cells in the originating network (or external data). These are arbitrary identifiers that will be remapped internally to sequential drive cell IDs (0 to n-1) in the target network. Different GIDs should be used for different source cells.
- locationstr
Target location of synapses in the target network. Must be ‘proximal’, ‘distal’, or ‘soma’, or a specific section name (when legacy_mode=False).
- weights_ampadict or None
Synaptic weights (in uS) of AMPA receptors for each targeted cell type (dict keys). Cell types omitted are set to zero.
- weights_nmdadict or None
Synaptic weights (in uS) of NMDA receptors for each targeted cell type (dict keys). Cell types omitted are set to zero.
- synaptic_delaysdict or float
Synaptic delay (in ms) at the column origin, dispersed laterally as a function of the space_constant. If float, applies to all target cell types. Use dict to create delay->cell mapping.
- space_constantfloat
Lateral dispersion constant (in units of inplane_distance) for synaptic weights and delays within the target network. Default: 3.0
- probabilityfloat or dict
Connection probability between source and target cells. Default: 1.0 (all-to-all).
- conn_seedint
Optional seed for random number generator for connectivity (default: None).
- cell_specificbool
If True, enables cell-specific connectivity (e.g., for 1-to-1 mapping). Default: False.
- n_drive_cellsstr or int
Number of drive cells. Use ‘n_cells’ for 1-to-1 mapping with target cell types, or an integer for a fixed number. Default: None (inferred from spike_data).
- add_tonic_bias(*, cell_type=None, section='soma', bias_name='tonic', amplitude, t0=0, tstop=None)[source]#
Attaches parameters of tonic bias input for given cell types
- Parameters:
- cell_typesstr | None
The name of the cell type to add a tonic input. When supplied, a float value must be provided with the amplitude keyword. Valid inputs are those listed in net.cell_types.
- sectionstr
name of cell section the bias should be applied to. See net.cell_types[cell_type].sections.keys()
- bias_namestr
The name of the bias.
- amplitude: dict | float
A dictionary of cell type keys (str) to amplitude values (float). Valid inputs for cell types are those listed in net.cell_types. If cell_types is not None, amplitude should be a float indicating the amplitude of the tonic input for the specified cell type.
- t0float
The start time of tonic input (in ms). Default: 0 (beginning of simulation). This value will be applied to all the tonic biases if multiple are specified with the amplitude keyword.
- tstopfloat
The end time of tonic input (in ms). Default: end of simulation. This value will be applied to all the tonic biases if multiple are specified with the amplitude keyword.
- copy()[source]#
Return a copy of the Network instance
The returned copy retains the intrinsic connectivity between cells, as well as those of any external drives or biases added to the network. The parameters of drive dynamics are also retained, but the instantiated
eventsof the drives are cleared. This allows iterating over the values defining drive dynamics, without the need to re-define connectivity. Extracellular recording arrays are retained in the network, but cleared of existing data.- Returns:
- net_copyinstance of Network
A copy of the instance with previous simulation results and
eventsof external drives removed.
- get_global_synaptic_gains()[source]#
Retrieve gain values for different celltype connections in the Network.
This function identifies excitatory and inhibitory cells in the Network and retrieves the gain value for each type of synaptic connection:
excitatory to excitatory (e_e)
excitatory to inhibitory (e_i)
inhibitory to excitatory (i_e)
inhibitory to inhibitory (i_i)
The gain is assumed to be uniform across all instances of each connection type (for example, between AMPA and NMDA, and between L2_pyramidal->L2_pyramidal and L2_pyramidal->L5_pyramidal, etc.). Only the first connection’s gain value is used for each type.
This does not return the synaptic gains of external drives.
- Returns:
- valuesdict
A dictionary with the connection types (‘e_e’, ‘e_i’, ‘i_e’, ‘i_i’) as keys and their corresponding gain values.
- plot_cells(ax=None, show=True)[source]#
Plot the cells using Network.pos_dict.
- Parameters:
- axinstance of matplotlib Axes3D | None
An axis object from matplotlib. If None, a new figure is created.
- showbool
If True, show the figure.
- Returns:
- figinstance of matplotlib Figure
The matplotlib figure handle.
- set_cell_positions(*, inplane_distance=None, layer_separation=None)[source]#
Set relative positions of cells arranged in a square grid
Note that it is possible to change only a subset of the parameters (the default value of each is None, which implies no change).
- Parameters:
- inplane_distancefloat
The in plane-distance (in um) between pyramidal cell somas in the square grid. Note that this parameter does not affect the amplitude of the dipole moment.
- layer_separationfloat
The separation of pyramidal cell soma layers 2/3 and 5. Note that this parameter does not affect the amplitude of the dipole moment.
- set_global_synaptic_gains(e_e=None, e_i=None, i_e=None, i_i=None, copy=False)[source]#
Change the synaptic gains of the celltypes in the Network.
- Parameters:
- e_efloat, default=None
Synaptic gain of excitatory to excitatory connections
- e_ifloat, default=None
Synaptic gain of excitatory to inhibitory connections
- i_efloat, default=None
Synaptic gain of inhibitory to excitatory connections
- i_ifloat, default=None
Synaptic gain of inhibitory to inhibitory connections
- copybool, default=False
If True, returns a copy of the network. If False, the network is updated in place with a return of None.
- Returns:
- netinstance of Network
A copy of the instance with updated synaptic gains if copy=True.
Notes
Synaptic gains must be non-negative. The synaptic gains will only be updated if a float value is provided. If None is provided (the default), the synaptic gain will remain unchanged.
This does not change the synaptic gains of external drives.
- write_configuration(fname, overwrite=True)[source]#
Writes network configuration to a json file.
Writes network configurations as a hierarchical json similar to the Network object’s structure. Outputs recorded during simulation such as currents and voltages are not saved due to size.
- Parameters:
- netNetwork
hnn-core Network object
- outputstr, Path, or StringIO
Path or buffer to write outputs
- overwritebool
Overwrite file if it exists. Default: True
- Returns:
- None